14 November 2025 | Friday | News
Streamlined, high-throughput sequencing of Influenza A virus with integrated indexing for Oxford Nanopore Technologies® platforms
New England Biolabs (NEB®) announces the release of the NEBNext Flu A Integrated Indexing Primer Module (NEB #E3436), designed to streamline and scale the sequencing of Influenza A virus from a range of species and sample types using Oxford Nanopore Technologies (ONT) platforms. This launch marks a significant step forward in enabling rapid, high-throughput surveillance of seasonal and emerging flu subtypes (e.g., H3N2, H1N1, and H5N1).
Developed in collaboration with the Johns Hopkins Applied Physics Laboratory (APL), a university affiliated research center advancing science and technology for global health resilience and security, the primer module supports a multi-segment RT-PCR workflow with integrated indexing ("iiMS-PCR"), allowing up to 48 samples to be sequenced in a run. The module is optimized for use in surveillance of zoonotic pathogens, public health labs, academic research and wastewater-based epidemiology programs.
"To effectively limit the spread of emerging biological threats—such as highly pathogenic avian influenza—genetic characterization methods must become as fast and simple to perform as diagnostic assays. Streamlined approaches to enable low-cost, high-throughput whole-genome sequencing support rapid local response efforts, ensuring that genomic data can be quickly integrated into regional and global monitoring systems to strengthen predictive modeling and preparedness capabilities," said Peter Thielen, a molecular biologist at APL.
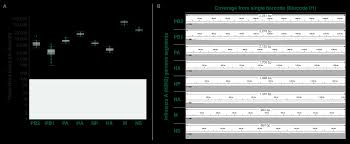
According to Dr. Erik Karlsson, Head of the Virology Unit at Institut Pasteur du Cambodge, "The iiMS workflow has been pivotal for rapid response to H5 outbreaks in both poultry and humans. It enables us to confirm poultry-to-human transmission within 24 hours, which is essential for timely public-health action and risk communication. We also rely on this platform weekly for routine assessment of circulating influenza strains across Cambodia." He adds, "The simplification of the workflow translates directly to rapid establishment in low-resource settings, allowing surveillance to happen where it is needed most and ensuring advanced genomic tools are available at the true frontlines of emerging zoonotic threats."
The NEBNext Flu A Integrated Indexing Primer Module includes reagents for indexed cDNA synthesis, amplification, and library preparation, and it works seamlessly with NEB's Monarch® Mag Viral DNA/RNA Extraction Kit, LunaScript® Multiplex One-Step RT-PCR Kit, and ONT's Native Barcoding Workflow. The protocol was proactively released on protocols.io to enable early access to this helpful workflow and has since been validated in diverse sample types, including clinical samples, agricultural products, and wastewater.
"Influenza A remains a persistent global health challenge due to its rapid evolution and seasonal variability," said Betsy Young, Ph.D., Senior Product Marketing Manager for Next Generation Sequencing at NEB. "With this primer module, we're equipping researchers and surveillance teams with a robust, scalable workflow to track genetic changes and inform deployment of public health resources."
The release of this product expands NEB's portfolio of reagents that support infectious disease research, and complements NEB's existing offerings for SARS-CoV-2, RSV, and other respiratory pathogens.
© 2026 Biopharma Boardroom. All Rights Reserved.