12 June 2024 | Wednesday | News
Illumina Inc. (NASDAQ: ILMN), a global leader in DNA sequencing and array-based technologies, today announced the launch of DRAGEN™ v4.3, the latest version of its DRAGEN™ software, part of the Illumina Connected Software portfolio, for analysis of next-generation sequencing data.
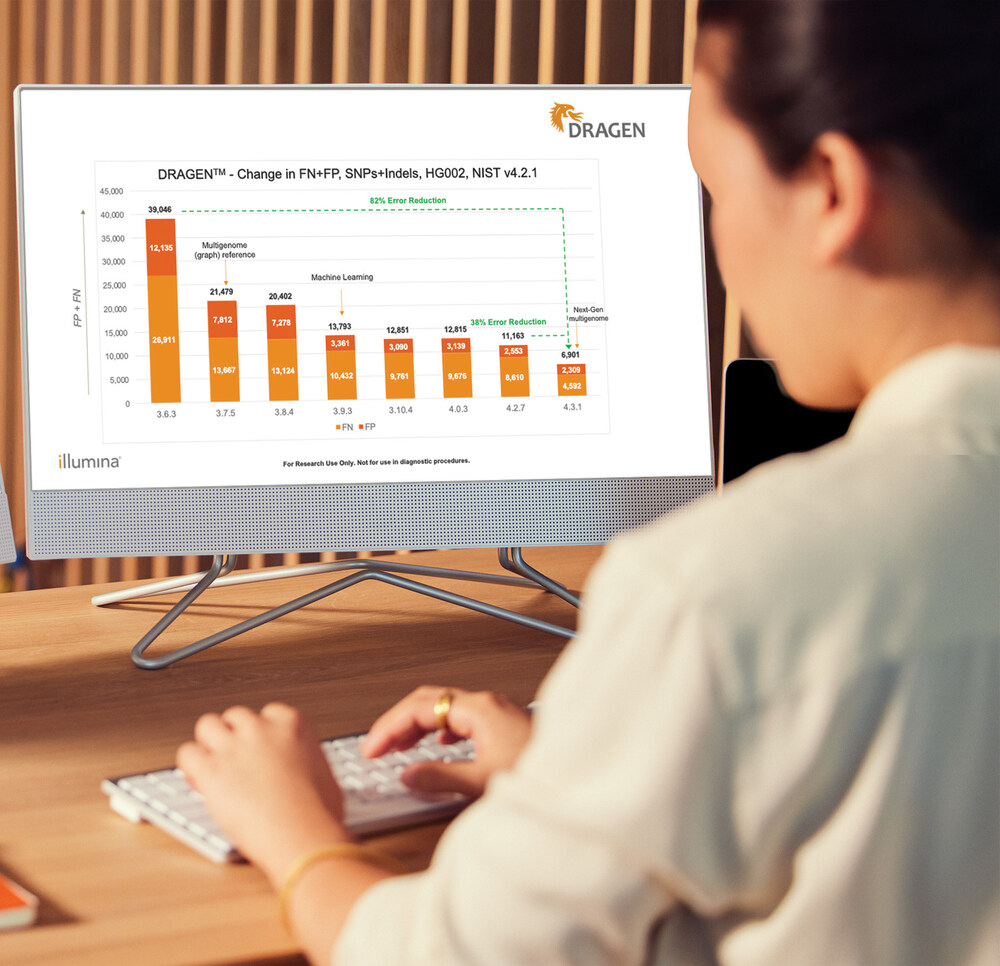
"We are excited to launch DRAGEN v4.3, with significant enhancements and additions to our analysis tools, empowering researchers and geneticists to maximize the value of the genome," said Rami Mehio, head of Software & Informatics at Illumina. "DRAGEN v4.3 includes our next-generation and most accurate multigenome mapping technology, built-in mosaic calling, advancements in machine learning, and the ability to genotype difficult genes to unlock deeper insights into the human genome with greater power, accuracy, and efficiency."
DRAGEN's innovation in multigenome mapping complements the efforts led by the Human Pangenome Reference Consortium (HPRC). The HPRC, funded by the National Human Genome Research Institute, aims to equip the genomics community with high-quality assemblies from diverse ancestries and combine them into a reference human pangenome.
"We have already delivered 47 high-quality assemblies that are now used by researchers around the globe," said Karen Miga, PhD, director of the Reference Production Center at the HPRC and assistant professor, Biomolecular Engineering, University of California, Santa Cruz. "With DRAGEN v4.3, and through the cloud apps, a DRAGEN customer can now leverage all or a subset of the 47 HPRC samples, allowing the user to select the ancestries of choice to power their DRAGEN multigenome mapping."
Benedict Paten, PhD, co-lead of the HPRC and associate professor, Biomolecular Engineering, University of California, Santa Cruz, added, "The accuracy improvement over traditional mapping is substantial and is expected to continue to improve when additional assemblies are incorporated in the pangenome."
DRAGEN v4.3 features a host of industry-leading innovations that together enable a more comprehensive genome, including:
Broad Clinical Labs has been systematically researching the accuracy of variant calls made by DRAGEN v4.3. Marina DiStefano, PhD, FACMG, associate lab director at Broad Clinical Labs, said: "We generated whole-genome sequence data from samples with confirmed calls in genes covered by DRAGEN v4.3 specialized callers, such as PMS2, NEB, and HBA, and from samples with orthogonally confirmed CNV calls. We also analyzed very-high-coverage WGS samples for mosaic evaluation. We are happy with the accuracy and speed and believe that DRAGEN v4.3 is a powerful tool for our clinical whole-genome research."
© 2026 Biopharma Boardroom. All Rights Reserved.